Software
The AI4Science group develops software for the simulation of molecular processes.
Our principles for scientific software development are
- clean design and reusable code
- ensuring correct functionality by unit and integration tests
- extensive documentation and sample notebooks
- automatic testing and deployment of each release on all target platforms
- open-source policy and development via Github
- workshops to provide hands-on training to users
We believe that the benefits of this effort outweigh the time and costs involved to maintain it: Software development and dissemination will increase the impact of developed methods, help to maintain group knowledge, serve the community and ensure reproducibility of results.
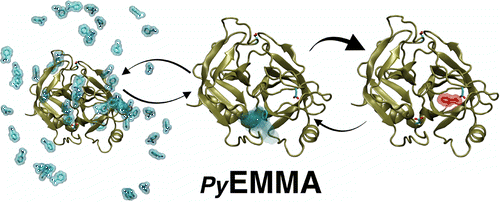
PyEMMA is a Python library for the estimation, validation, and analysis of kinetic models from MD data. Functionalities include dimension reduction techniques such as the time-lagged independent component analysis, clustering, maximum-likelihood and Bayesian estimation of Markov State Models and Hidden Markov Models, coarse-graining and analysis of kinetic models, computation of transition pathways, and visualization tools.
Code: github.com/markovmodel/pyemma
Languages: Python (90%), C (10%)
Usage: > 50,000 downloads
License: Lesser GPL (open-source)
Release: 2.4, 34 releases
Platforms: Linux, Windows, Mac
DOI: 10.1021/acs.jctc.5b00743
DOI: 10.1021/ct300274u
The one-week PyEMMA winter school in February is open to students and researchers.
For more detail see past winter school programs and our youtube channel.
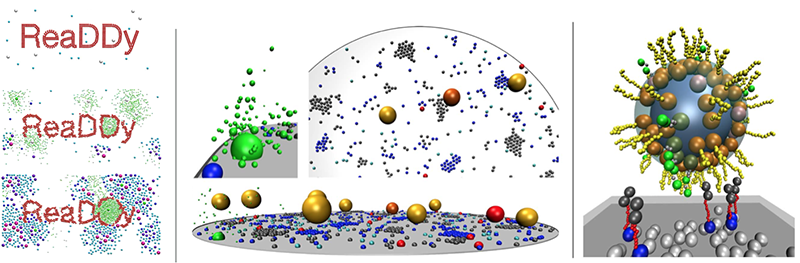
ReaDDy: High-performance simulator for reaction-diffusion dynamics with interacting particles. Suitable for modeling cellular signal transduction, chemical microreactors, soft matter systems, membrane mechanics, virus assembly etc. Previous version was Java-based. New version currently developed with software abstraction layers: (i) Python user API, (ii) C++ API, (iii) Multithreading/CUDA/MPI kernels for execution of different platforms.
Code: github.com/readdy/readdy
Languages: C++ (70%) CUDA (20%) Python (10%)
Usage: ca. 1,000 downloads
License: Lesser GPL (open-source)
Releases: 1.0 (Java), v1.0.0 (C++)
Platforms: Linux, Mac
DOI: 10.1371/journal.pone.0074261
DOI: 10.1016/j.bpj.2014.12.025
Our 1-week ReaDDy summer school in July is open to students and researchers. For information on the upcoming summer school see here.
Library Markov chain inference Python/C, LGPL license. v1.2.1 (12 releases).
Library for Hidden Markov model inference Python/C, LGPL license. v0.5.2 (9 releases).
Library for statistical inference from multi-ensemble data, Python/C, LGPL license. v0.2.6 (22 releases).
Python module for interactive visualization of projected coordinates of molecular dynamics (MD) trajectories.
Deep learning meets molecular dynamics. In development.