You are here: Foswiki>ABI Web>LectureWiki>PMSB_Seqan_2016>PSMB_Seqan_2016_p4 (28 Jan 2016, h4nn3s)Edit Attach
PSMB_Seqan_2016_p4
In this project you will implement the typical Needleman-Wunsch-Algorithm, but with a few twists that will hopefully allow "auto-vectorization"Introduction
The classical Needleman-Wunsch algorithm is one of the most famous algorithms in bioinformatics and you have likely studied it before. One of the heuristic modifications to it, is to use a so called band, that reduces the computational space and time from quadratic to linear. Independent of this modification the cells in the dynamic programming matrix are computed in column-wise (or row-wise). In this order all cells in one row are dependent on one another which prevents "vectorization", a feature of modern processors that can greatly speed up different kinds of computations. To remedy this, you will instead compute the cells anti-diagonal-wise: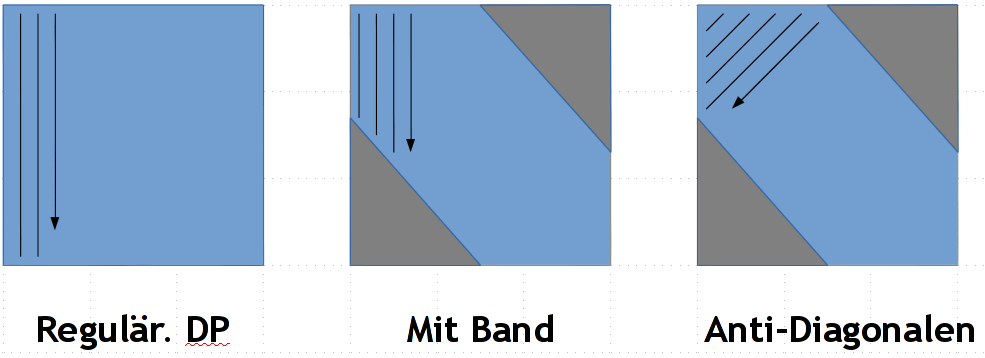
Tasks
- read the literature about the algorithm and about vectorization
- Implement the algorithm as described above
- see if auto-vectorization works, play with OpenMP if it doesn't
- compare to existing SeqAn implementation (without vectorization)
Stretch goals
- use affine gap costs
- use scoring matrices, make sure amino-acid-sequences work as well
Extension as Bachelor Project
- cover more algorithmic combinations, also Smith-Waterman
- also alignment extension
- integrate with SeqAn
- write documentation
Literature
TODO needleman wunsch; banded modification; anti-diagonal paperEdit | Attach | Print version | History: r3 < r2 < r1 | Backlinks | View wiki text | Edit wiki text | More topic actions
Topic revision: r2 - 28 Jan 2016, h4nn3s
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Foswiki? Send feedback
