Page AdaptiveMolecularDynamicsSimulation
The main page about Adaptive Molecular Dynamics Simulation (AMoDS)Project Overview
The omnipresent Sampling Problem is one of the big issues in Molecular Dynamics Simulation. So far the amount of information, that needs to be computed is too large, so that one cannot simulate processes of timescales of general interest. This means, that we cannot circumvent the Sampling Problem, the only ways we can deal with this is to either increase the computational effort or simulate at some degree of approximation.A third not often taken option is to restrain the simulation not to the full information contained in the phase-space, but concentrate the simulation on a certain kind and leave all other information aside. In every case a certain amount of information costs a certain amount of computational ressources, we will follow the third option and try to intelligently design a set of short simulations, that will give a maximum of information about a specific, preselected information about the system. This, of course, has the drawback, that the gained information might be of lesser use for other problems.
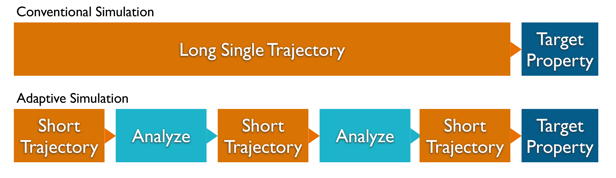
To achieve a guidance of the simulation without interfering or biasing, we divide the available simulation into chunks of short trajectories (s. \prettyref{fig:enhancedSampling}). Each of this simulation parameters are chosen carefully, to maximize the information in the target property.
The Adaptive Cycle
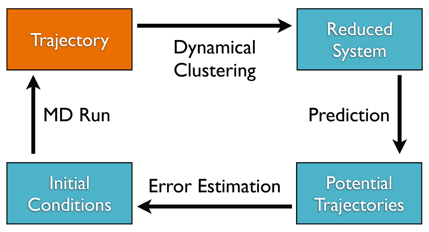
The Adaptive Cycle consists of four steps, that are applied until a certain amount of convergence of the target property is achieved. Given a first trajectory as input :
- Clustering of the given trajectory into metastable states
- Prediction of Transition Matrices depending on various initial condition and information from previous trajectory
- Computing of Errors in the target property for each Prediction
- Running a simulation with the initial conditons, that reduce the error most
Related Links
kommen noch
Comments
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Foswiki? Send feedback
